Orthology Basics
Differents types of homologs in OMA
In OMA, we infer and provide several sub-types of orthologs: pairwise orthologs, Hierarchical Orthologous Groups (HOGs), and OMA Groups . It is important to understand how these three categories of orthologs are different in order to choose the appropriate type for your analysis. The differences between the three sub-types of orthologs reported in OMA are all based on one main factor: how they are inferred. In the OMA algorithm, the pairwise orthologs are inferred first, and are then used to build the HOGs and OMA Groups.
Pairwise Orthologs
- Pairs of orthologs are inferred based on the sequence similarity of genes between genomes.
- In OMA, we also report the relationship cardinality of the pairwise orthologs, which reflects the level of co-orthology, or the degree of duplications which one or both of the orthologs has undergone. One-to-one (1:1) pairwise orthology means that both genes in the pair have only one ortholog in the other species. A one-to-many relationship (1:m) means that the gene of interest has more than one ortholog in the other species. This implies that the gene was duplicated in an ancestor of the other species, but after the speciation event. A many-to-many (m:m) relationship means both orthologs underwent lineage-specific duplications.
- For how to find pairwise orthologs in the OMA browser, see Access the OMA DataーOrthologs of a given gene .
Hierarchical Orthologous Groups (HOGs)
- HOGs aim to identify sets of genes that have descended from a common ancestral gene in a given ancestral species (i.e. at a specific taxonomic level).
- The pairwise orthologs are mapped to an orthology graph, where each node on the graph represents a gene, and each solid line between the genes represents an inferred pairwise orthologous relationship. The graphs are then used as input to compute the HOGs.
- HOGs are constructed by identifying groups in the graph of pairwise orthologs. Each of the connected components in the graph are putative gene families, composed of genes which descended from a common ancestral gene.
- After forming the groups of HOGs, HOG-derived orthologs can be considered as any pairs of genes between species which are contained in the same HOG, given that the HOG is defined at the level of their last common ancestor.
- The “hierarchical” nature of HOGs is because they are defined with respect to specific taxonomic clades. Groups defined at more recent clades are encompassed within larger groups that are defined at older clades, thus making them nested subfamilies.
- For how to find HOGs in OMA, see Access the OMA Data .
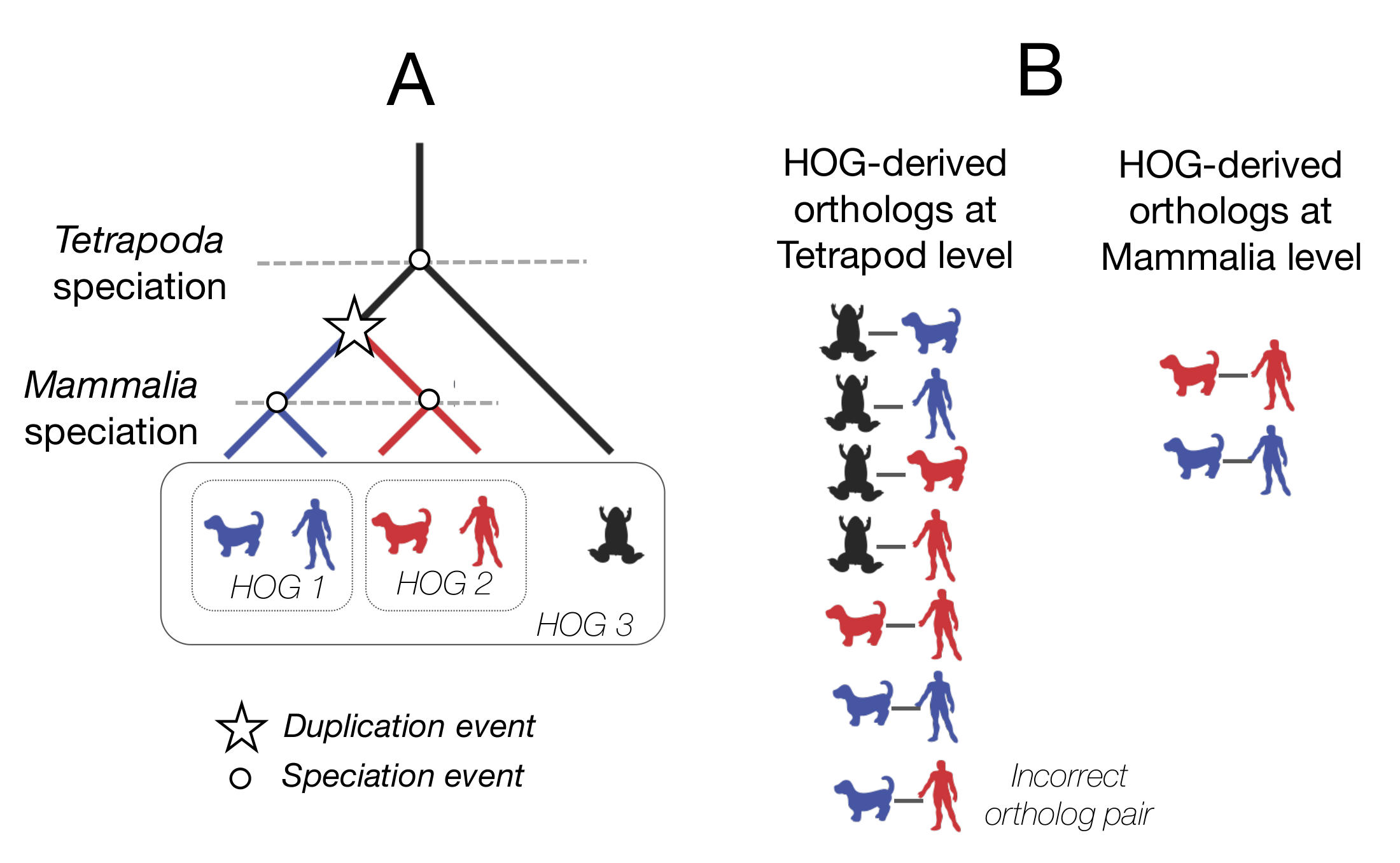
An example of the hierarchical nature of HOGs. At the mammalian level, there are two separate HOGs. At the tetrapod level, there is 1 HOG, encompassing the smaller, mammalian HOGs.
OMA Groups
- OMA Groups are cliques of orthologs based on the orthology graph. In mathematics, a clique is a part of a graph where each node in that part is connected to all other nodes in that same part. Thus, in an OMA Group, all the genes are connected to each other by pairwise orthologous relations.
- While pairwise orthologs and hierarchical orthologous groups are commonly used terms, OMA Groups is a term specific to OMA. However, sometimes they are referred to as “Orthologous Groups” (see (Dylus et al. 2020)).
- One common misconception is that OMA Groups are groups of 1:1 orthologs. This is not necessarily the case. If two genes are 1:1 orthologs, this implies that there are no co-orthologs of either gene. If co-orthologs are inferred, OMA Groups will only contain one of the co-orthologous copies. However, like sets of 1:1 orthologs, OMA Groups have the property that all members are orthologous to all other members of the same group.
- For how to find OMA Groups in OMA, see Access the OMA Data .
Paralogs
- Paralogs in OMA are derived from the HOGs. That is, they originated from an inferred duplication event.
- For how to find paralogs in OMA, see Access the OMA Data .